Generating Molecules using CharRNN
In this blog we are going to discuss about RNN’s and How the character RNN algorithms can be used in Molecular Structure Generation.
Before we go deeper into the details of Recurrent Neural networks, lets just go through some methods and procedures about RNN. Note: When you are going through the article,it is important for you to have basic knowledge of neural networks and how Pytorch (a deep learning library) works. You can also refer to some other articles to understand these concepts in broader sense: In this post ,I am implementing a RNN model with Pytorch to generate SMILES.
Now in this we will learn-
- Why/what are Recurrent Neural Networks?
- Character level RNN model
- RNN for Molecules (SMILES) Generation
- Generating SMILES using RNN's
Recurrent Neural Networks-The idea behind RNN’s is to make use of sequential information.Here the order of the data plays an important role, like audio or sentences. What makes Recurrent Networks so special? They allow us to operate over sequences of vectors and this thing gives importance:
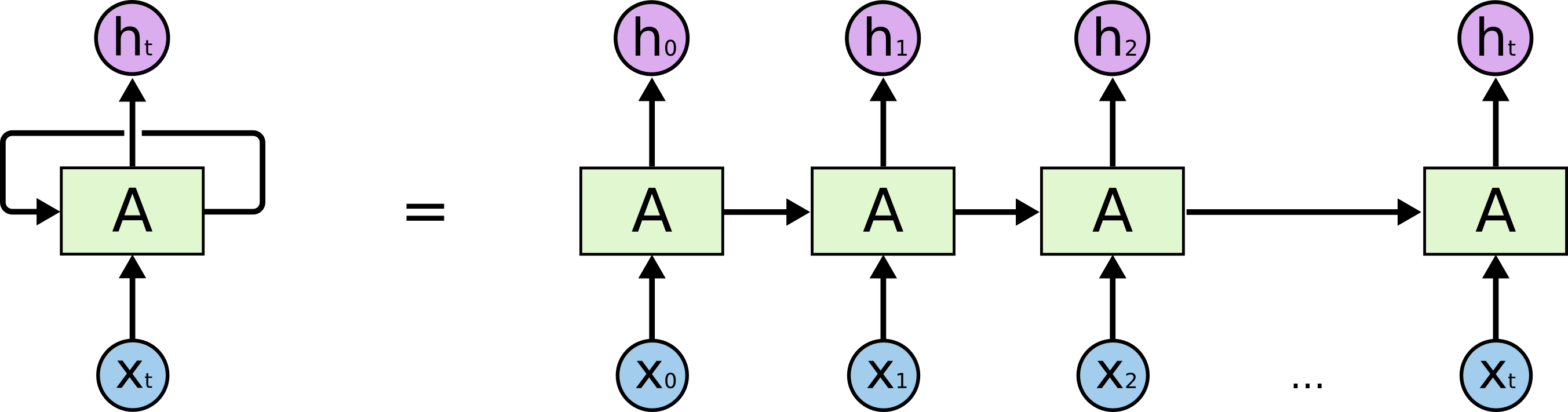
Character-Level RNN Model: Okay, so we have an idea about what RNNs are, why they are super exciting, and how they work. We’ll now ground this in a fun application: We’ll train RNN character-level RNN models. That is, we’ll give the RNN a huge chunk of data(Smiles representation of molecules)and ask it to model the probability distribution of the next character in the sequence given a sequence of previous characters. This will only allow us to generate new smiles and one character at a time. By the way, together with this post I am also releasing (code on Github:Visit this link) that allows you to train char RNN model based on multi-layer LSTMs.
RNN for Molecules (SMILES) Generation- In this Post, we want to show that recurrent neural networks can be trained as generative models for molecular structures. It is similar to statistical language models in natural language processing. We demonstrate that the properties of the generated molecules correlate very well with the properties of the molecules used to train the model. To connect chemistry with language, it is important to understand how molecules are represented. Usually, they are modeled by molecular graphs, also called Lewis structures in chemistry. In molecular graphs, atoms are labeled nodes. The edges are the bonds between atoms, which are labeled with the bond order (e.g., single, double, or triple).
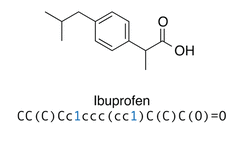
However, in models for natural language processing, the input and output of the model are usually sequences of single letters, strings or words.Therefore we employ the SMILES (Simplified Molecular Input Line Entry System) format.This pattern are the type of chemical notation that helps us to represent molecules. They can also be used by computers easily. It is a simple string representation of molecules which encodes molecular graphs compactly as human-readable strings. SMILES is a formal grammar which describes molecules with an alphabet of characters, for example c and C for aromatic and aliphatic carbon atoms, O for oxygen, and −, =, and # for single, double, and triple bonds (see Figure 2).To indicate rings, a number is introduced at the two atoms where the ring is closed. For example, benzene in aromatic SMILES notation would be c1ccccc1.
Generating SMILES using RNN’s: I’ll be showing you how I implemented my recurrent neural network in Pytorch. I trained it using the ChEMBL smiles Dataset ,which contains 2M smiles and it is a manually curated database of bio-active drug-like molecules.
Part 1: Importing libraries and data preprocessing - First, we import pytorch, the deep learning library we’ll be using.Also import nn (pytorch’s neural network library) and torch.nn.functional, which includes non-linear functions like ReLu and sigmoid.
Let’s load the Data file and name it as text
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
with open('chembl_smiles.txt ', 'r') as f:
text = f.read()
Then we’ll create a dictionary out of all the characters and map them to an integer. This will allow us to convert our input characters to their respective integers (char2int) and vice versa (int2char).
chars = tuple(set(text))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# Encode the text
encoded = np.array([char2int[ch] for ch in text])
Finally, we’re going to convert all the integers into one-hot vectors.
# Defining method to encode one hot labels
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
We will usually want to feed training data in batches to speed up the training process,so defining method to make mini-batches for training.
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
batch_size_total = batch_size * seq_length
# total number of batches we can make
n_batches = len(arr)//batch_size_total
# Keep only enough characters to make full batches
arr = arr[:n_batches * batch_size_total]
# Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
# iterate through the array, one sequence at a time
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n:n+seq_length]
# The targets, shifted by one
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, n+seq_length]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, 0]
yield x, y
Part 2: Building the Model First, we’re going to check if we can train using the GPU, which will make the training process much quicker. If you don’t have a GPU, be ready to witness the more time taken by it. Check out Google Collaboratory or other cloud computing services!
# Check if GPU is available
train_on_gpu = torch.cuda.is_available()
if(train_on_gpu):
print('Training on GPU!')
else:
print('No GPU available, training on CPU; consider making n_epochs very small.')
Now, we define our Char-RNN Model! .We will implement dropout for regularization and here rather than having the input sequence be in words, we’re going to look at the individual letters/characters instead.
# Declaring the model
class CharRNN(nn.Module):
def __init__(self, tokens, n_hidden=10, n_layers=2,
drop_prob=0.2, lr=0.001):# n_hidden=256,
super().__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
# creating character dictionaries
self.chars = tokens
self.int2char = dict(enumerate(self.chars))
self.char2int = {ch: ii for ii, ch in self.int2char.items()}
#define the LSTM
self.lstm = nn.LSTM(len(self.chars), n_hidden, n_layers,
dropout=drop_prob, batch_first=True)
#define a dropout layer
self.dropout = nn.Dropout(drop_prob)
#define the final, fully-connected output layer
self.fc = nn.Linear(n_hidden, len(self.chars))
For our forward function, we’ll propagate the input and memory values through the LSTM layer to get the output and next memory values. After performing dropout, we’ll reshape the output value to make it the proper dimensions for the fully connected layer.
def forward(self, x, hidden):
''' Forward pass through the network.
These inputs are x, and the hidden/cell state `hidden`. '''
#get the outputs and the new hidden state from the lstm
r_output, hidden = self.lstm(x, hidden)
#pass through a dropout layer
out = self.dropout(r_output)
# Stack up LSTM outputs using view
out = out.contiguous().view(-1, self.n_hidden)
#put x through the fully-connected layer
out = self.fc(out)
# return the final output and the hidden state
return out, hidden
Finally, for initializing the hidden value for the correct batch size if you’re using mini-batches.This method generates the first hidden state of zeros which we will use in the forward pass,We will send the tensor holding the hidden state to the device we specified earlier as well.
def init_hidden(self, batch_size):
''' Initializes hidden state '''
# Create two new tensors with sizes n_layers x batch_size x n_hidden,
#initialized to zero,for hidden state and cell state of LSTM
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda())
else:
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_())
return hidden
Part 3: We’ll declare a function, where we’ll define an optimizer(Adam) and loss (cross entropy loss). We will then proceed with the training and validation data and initialize the hidden state of the RNN. We’ll loop over the training set, each time encoding the data into one-hot vectors.We will perform forwarding and backpropagation and update the gradients. Please for full closure of the code, visit our Github profile(please for full code visit our Github profile)
We’ll have the method generate some loss statistics(training loss and validation loss) to let us know if the model is training correctly. Now, we’ll just declare the hyper parameters for our model, create an instance for it, and train it!
n_hidden=56
n_layers=2
net = CharRNN(chars, n_hidden, n_layers)print(net)
# Declaring the hyperparameters
batch_size = 32
seq_length = 50
n_epochs = 1
# start smaller if you are just testing initial behavior
train the modeltrain(net, encoded, epochs=n_epochs, batch_size=batch_size, seq_length=seq_length, lr=0.001,print_every=10000)
Part 4:The prediction task The input to the model will be a sequence of characters(smiles), and we train the model to predict the output - Since RNN’s maintain an internal state that depends on the previously seen elements, given all the characters computed until this moment, what is the next character? After training, we’ll create a method (function) to predict the next character from the trained RNN with forward propagation.’’’ Given a character, predict the next character. Returns the predicted character and the hidden state.
def predict(net, char, h=None, top_k=None):
# tensor inputs
x = np.array([[net.char2int[char]]])
x = one_hot_encode(x, len(net.chars))
inputs = torch.from_numpy(x)
if(train_on_gpu):
inputs = inputs.cuda()
# detach hidden state from history
h = tuple([each.data for each in h])
# get the output of the model
out, h = net(inputs, h)
# get the character probabilities
p = F.softmax(out, dim=1).data
if(train_on_gpu):
p = p.cpu() # move to cpu
# get top characters
if top_k is None:
top_ch = np.arange(len(net.chars))
else:
p, top_ch = p.topk(top_k)
top_ch = top_ch.numpy().squeeze()
# select the likely next character with some element of randomness
p = p.numpy().squeeze()
char = np.random.choice(top_ch, p=p/p.sum())
# return the encoded value of the predicted char and the hidden state
return net.int2char[char], h
Then, we’ll define a sampling method that will use the previous method to generate an entire string of smiles, first using the characters in the first word (prime) and then using a loop to generate the next words using the top_kfunction, which chooses the letter with the highest probability to be next.
def sample(net, size, prime='The', top_k=None):
if(train_on_gpu):
net.cuda()
else:
net.cpu()
net.eval() # eval mode
# First off, run through the prime characters
chars = [ch for ch in prime]
h = net.init_hidden(1)
for ch in prime:
char, h = predict(net, ch, h, top_k=top_k)
chars.append(char)
# Now pass in the previous character and get a new one
for ii in range(size):
char, h = predict(net, chars[-1], h, top_k=top_k)
chars.append(char)
return ''.join(chars)
# Generating new text
print(sample(net, 120, prime='A', top_k=5))
Finally, we just call the method, define the size you want (I chose 120 characters) and the prime (I chose ‘A’) and get the result!
Final Output-
A[N+](=O)[O-])[O-].Cl
CCCOC(=O)C(NCc1ccc(F)cc1)c1ccccc1
Cc1nc2ccc(Cl)cc2c2c(c2c1OCC(NC(=O)CNCc1ccccc1)CC1)CC2
COc1cc(C)cc2n(CCCNCC(=O)Nc3cc(C)nc(-c4cc(F
I hope you enjoyed coding up with molecular Generation!