Sequence-based ab initio prediction of variant effects on expression and disease risk using ExPecto
Understading disease from the DNA mutations is one of the major problem researchers are trying to solve. In this blog, we will analyze about a machine learning model which can predict disease risk from the sequence variation.
Introductory note:
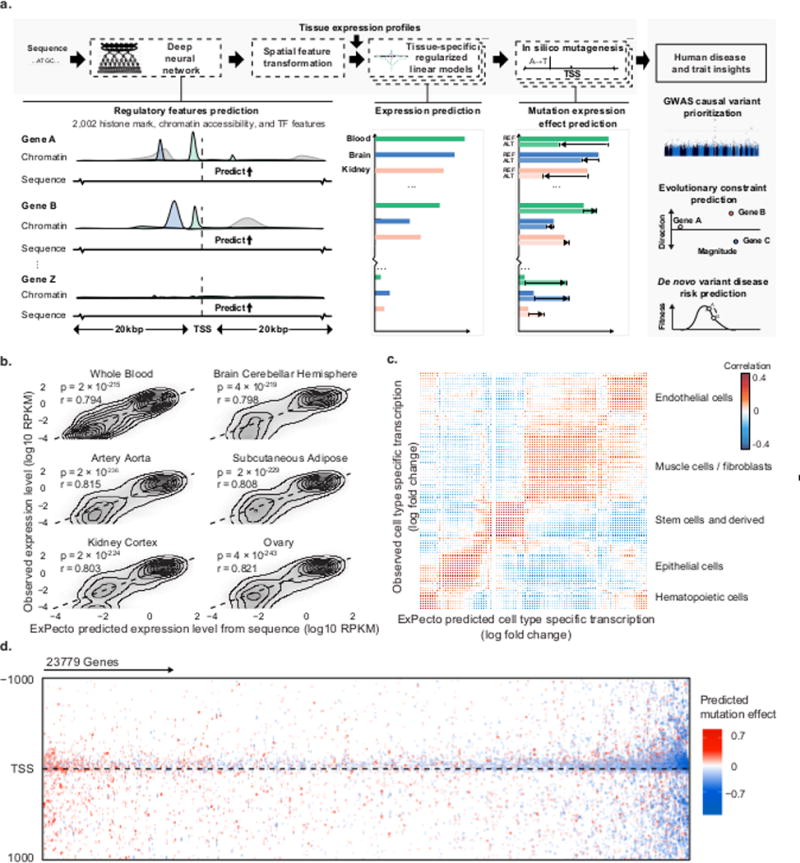
Human DNA sequence consists of genes and regulatory sequences. Genes are used for making proteins while regulatory sequences, also known as non-coding variant act as a regulatory body which decides many genomic functions. The function includes the time utilized in creating the protein, the quantity required for its formation. Today, the biggest challenge in human genetics is whether precision medicine and evolutionary biology are deciphering the regulatory code of gene expression. It also includes understanding the transcriptional effects of genome variation. To cope with this complication, we have used deep learning-based framework known as ExPecto. ExPecto can predict ab initio from the DNA sequence, the tissue-specific transcriptional effects of mutations, including rare or never observed.
ExPecto:
ExPecto model takes DNA sequence as input and predicts the future risk of a person from a disease. It uses a deep learning network model followed by spatial transformation and finally, it makes a tissue-specific prediction using L2-regularized linear regression models. The model is fitted by a gradient boosting algorithm. Thus, after getting a tissue-specific prediction, we infer the risk of disease which the patient may have in the future.
Model planning:
Input:
ExPecto takes a DNA sequence of length 40,000. We find the position of allele and then take the DNA sequence of length 20,000 on both left and right side. If we take less than 40k size, the performance decreases slightly. But if we increase the size and it is more than 40k, there is an insignificant chance of high performance. You will witness that in the below figure.
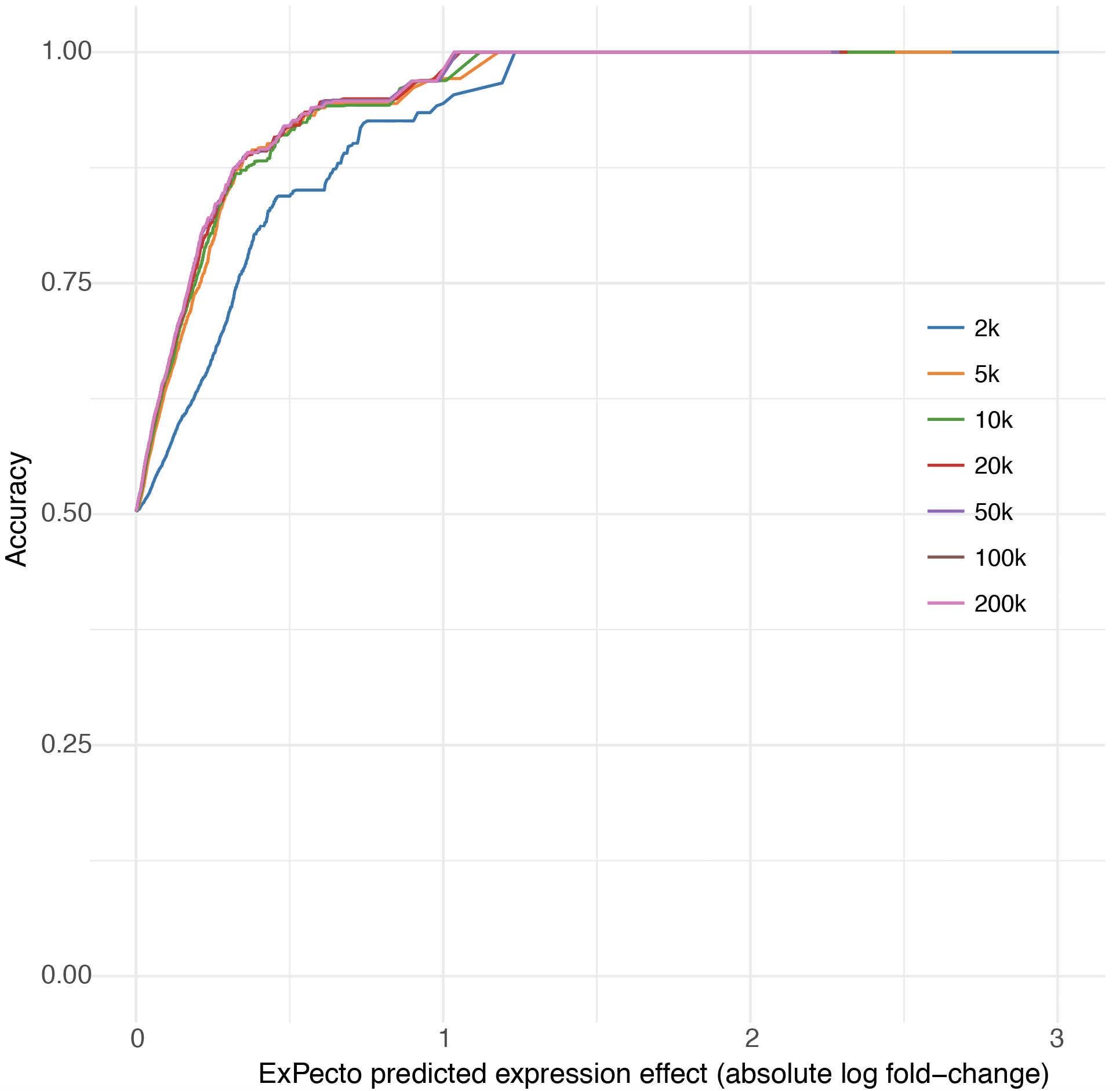
In the above figure, each line shows performance for the model trained with different window sizes (+/- around the TSS). The x-axis represents the predicted effect magnitude cutoff, as measured by absolute log fold-change. Y-axis represents the accuracy of predicting the expression change directionality for the variants above the corresponding effect magnitude.
Architecture:
ExPecto model consist of three components that act sequentially.
1) The deep convolutional network regulatory feature representation model architecture:
In total, we have 2002 genome-wide histone marks, transcription factor binding and chromatin accessibility features. Our deep learning model scans the sequences each of the length 2000. The deep convolutional neural network model predicts epigenomic features of a 200bp region. It also uses the 1800bp surrounding context sequence. Thus each 200 bp region will give 2002 features, thus ranging in a total of 400400 features.
Following is the architecture of deep convolutional network:
Input (Size: 4bases x 2000bp) =>
(Layer 1): Convolution(4 -> 320, kernel size=8)
(Layer 2): ReLU
(Layer 3): Convolution(320 -> 320, kernel size=8)
(Layer 4): ReLU
(Layer 5): Dropout(Probability=0.2)
(Layer 6): Max pooling(pooling size=4)
(Layer 7): Convolution(320 -> 480, kernel size=8)
(Layer 8): ReLU
(Layer 9): Convolution(480 -> 480, kernel size=8)
(Layer 10): ReLU
(Layer 11): Dropout(Probability=0.2)
(Layer 12): Max pooling(pooling size=4)
(Layer 13): Convolution(480 -> 960, kernel size=8)
(Layer 14): ReLU
(Layer 15): Convolution(960 -> 960, kernel size=8)
(Layer 16): ReLU
(Layer 17): Dropout(Probability=0.2)
(Layer 18): Linear(101760 -> 2003)
(Layer 19): ReLU
(Layer 20): Linear(2003 -> 2002)
(Layer 21): Sigmoid
=> Output (Size: 2002 epigenomic features)
ReLU indicates the rectified linear unit activation function. Sigmoid indicates the Sigmoid activation function. Notations such as 4 -> 320 indicate the input and output channel size for each layer.
2) Spatial Feature Transformation:
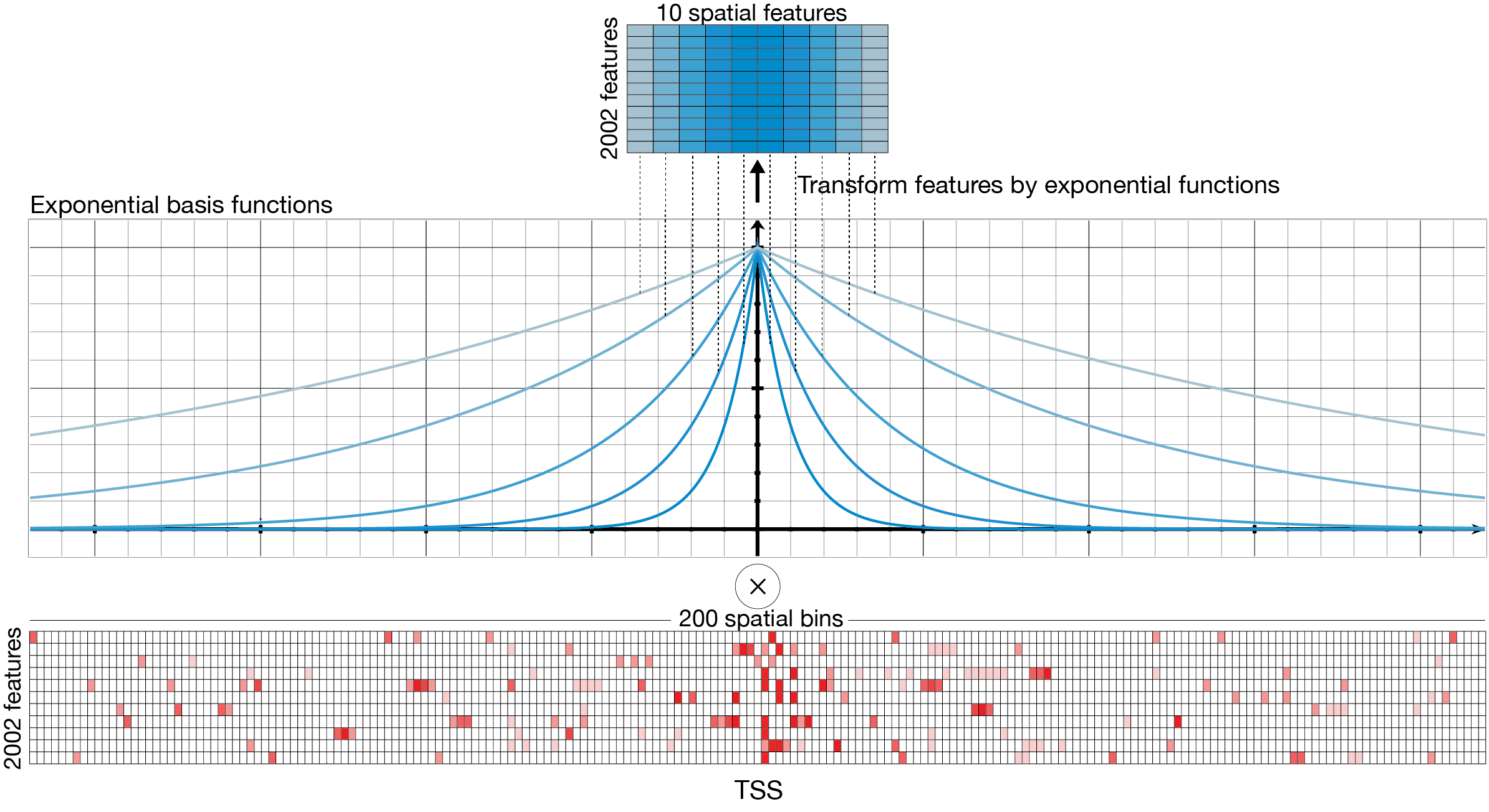
The second component of ExPecto is the spatial transformation module that reduces the dimensionality of the learning problem by generating spatially-transformed features. The spatial transformation module reduces the input dimensionality with ten exponential functions weighting upstream and downstream regions separately. The weights are based on the relative distance to TSS (transformed features with a higher decay rate are more concentrated near TSSs). This effectively reduces the number of features from 20 fold to 20020. The exponential functions were prespecified (based on empirical selection) and not learned during training. Scanning the 40kb region surrounding the TSS with the deep convolutional network model outputs 200 spatial bins for each of the 2002 histone mark, transcription factor, and DNase features. Spatial transformation condenses this information into 10 spatial features by using exponential functions with 5 different decay rates to compute the weighted sum over spatial bins. Upstream regions and downstream regions are transformed independently.
3) L2-regularized linear regression model:
Finally, to make tissue-specific expression predictions, spatially-transformed features are used to predict gene expression levels. For each tissue, L2-regularized linear regression models fitted by gradient boosting algorithm are used. Specifically, the full models including both spatial transformation and linear models are specified as below.
Summary:
So using this model we can predict the risk of disease in a person so that he can start with his treatment immediately.