Molecular Structure Generation using VAE
This post gives a detailed explanation about using Variational Autoencoders for Molecular structure Generation. By the end of this post, you will be able to create your VAE or molecular generation.
In this blog, we will be using Variational AutoEncoder for molecular structure generation.
If you don’t know about VAE, please visit VAE to understand it better.
Molecular generation is producing new molecules that have a valid structure. In this model, we generate a similar molecule as input molecules. The input of this model is smiles and the output is similar to that of smiles as of input. We use pytorch to implement VAE in a molecular generation.

SMILES (Simplified Molecular Input Line Entry System) are the type of chemical notation that helps us to represent molecules and easy to be used by the computers. It is a simple string representation of molecules. Some examples include
c1ccccc1 benzene,
c1c(N(=O)=O)cccc1 Nitrobenzene
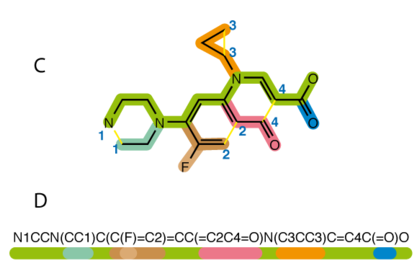
Here we see molecular generation in pytorch. For the implementation of VAE in the molecular generation, we will be using ChEMBL smiles dataset which contains 2M smiles, and it is a manually curated database of bioactive drug-like molecules.
We will also be using RNN in this model because RNN tries to capture the pattern of text easily as compared to CNN and also in RNN. We also use LSTM and GRU for memorizing.
We use the GRU (Gated Recurrent Unit) model because it aims in solving the vanishing gradient problem. It comes with a standard recurrent neural network.
Let’s now get our hands dirty with the code.
for full code click
Import dataset
the dataset we used for this purpose is refined from Zinc Database dataset
This is huge dataset so first slice the dataset according to your system.This dataset contain 1,936k smiles.
We use only 1k smiles for faster computation.
First, we will import our dataset, which contains smiles and convert into the strings.
import pandas as pd
import torch
data = pd.read_csv('.../dataset_iso_v1.csv')
train_data1 = data[data['SPLIT'] == 'train']
train_data_smiles2 = (train_data1["SMILES"].squeeze()).astype(str).tolist()
train_data = train_data_smiles2
Constructing a vocabulary
After that, we build a vocabulary for the model. In order to create the vocabulary, we use set() because when we feed data into the set, it removes the repeat data. After that, we make a list of characters to make vocabulary for our model then we add <’bos’> to indicate the beginning of smiles,<’eos’> to indicate the end of a sentence,<”pad”> to make all smiles of the same length and <”unk”> for unknown words.
After completion of vocabulary we make character to index and index to character, to encode decode the vocabulary.
To decrease the time for training the model, we use Cuda in pytorch. This is done for changing our computation from CPU to GPU because GPU handles lots of parallel computations using thousands of cores. Also, they have a large memory bandwidth to deal with the data for these computations.
After completion of vocabulary, we then feed it into the embedding matrix. But we will discuss embedding matrix later.
We will create several functions which we use later in the model character to ids, ids to character, the string to ids, ids to string, string to tensor. You can see the functions and you will understand the reason behind creating these functions.
chars = set()
for string in train_data:
chars.update(string)
all_sys = sorted(list(chars)) + ['<bos>', '<eos>', '<pad>', '<unk>']
vocab = all_sys
c2i = {c: i for i, c in enumerate(all_sys)}
i2c = {i: c for i, c in enumerate(all_sys)}
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
vector = torch.eye(len(c2i))
...
visit above link for full code
Model
In the previous post about VAE part, I have explained about the role of the encoder, decoder and latent vector.As we now create vae model, if you have doubts in the theory part of VAE, you can go through my previous post.
We feed our vocabulary and one hot matrix into the embedding matrix. Embedding is a way to represent a character or word into a dense vector representation. Embeddings are useful because they can reduce the dimensionality of categorical variables and meaningfully represent categories in the transformed space.
We then feed our embedding dimension into the encoder. The encoder encodes our input sample data into small vector from which we call latent vector. This small vector represents our entire dataset. The encoder output gives latent space vector and kl divergence loss The decoder work is to decode the latent space and bring back our input data. It is not necessary that our encoder and decoder remains the same. We can change the layers according to our want. The latent sample takes probability distributions of various characteristics of our input. Let’s say we have a latent vector of 3 nodes. This means our latent space vector defines our input into these 3 nodes and these 3 nodes contain a distribution of characteristics of the input dataset. We take a sample data from this distribution the whole code of VAE model is:
class VAE(nn.Module):
def __init__(self,vocab,vector):
super().__init__()
self.vocabulary = vocab
self.vector = vector
n_vocab, d_emb = len(vocab), vector.size(1)
self.x_emb = nn.Embedding(n_vocab, d_emb, c2i['<pad>'])
self.x_emb.weight.data.copy_(vector)
#ENCODER
self.encoder_rnn = nn.GRU(d_emb,q_d_h,num_layers=q_n_layers,batch_first=True,dropout=q_dropout if q_n_layers > 1 else 0,bidirectional=q_bidir)
q_d_last = q_d_h * (2 if q_bidir else 1)
self.q_mu = nn.Linear(q_d_last, d_z)
self.q_logvar = nn.Linear(q_d_last, d_z)
# Decoder
self.decoder_rnn = nn.GRU(d_emb + d_z,d_d_h,num_layers=d_n_layers,batch_first=True,dropout=d_dropout if d_n_layers > 1 else 0)
self.decoder_latent = nn.Linear(d_z, d_d_h)
self.decoder_fullyc = nn.Linear(d_d_h, n_vocab)
# Grouping the model's parameters
self.encoder = nn.ModuleList([self.encoder_rnn,self.q_mu,self.q_logvar])
self.decoder = nn.ModuleList([self.decoder_rnn,self.decoder_latent,self.decoder_fullyc])
self.vae = nn.ModuleList([self.x_emb,self.encoder,self.decoder])
..
for full code visit above link
Training
We use inbuilt pytorch function DataLoader to create batches and keep similar length smiles into the same batch. In data loader we have collate function which helps to convert data into tensors and do padding to make similar length data.it is not necessary to use this collate function if we have a same length of data or we convert our data into tensors already.
Then the data which comes from the dataloader are used by us in the training process. In this training, we use Adam optimizer for optimization, and then we train our data into 50 epoch(one epoch means the whole model run 1 time)
By using this, we try to minimize KL-divergence loss and Reconstruction loss. I have already explained both losses in my previous post.
Reconstruction loss is how well our model reconstructs back the input data and kl-divergence loss measure how much information is lost when using q to represent p. Remember these q and p is data distribution.
def _train_epoch(model, epoch, train_loader, kl_weight, optimizer=None):
if optimizer is None:
model.eval()
else:
model.train()
kl_loss_values = CircularBuffer(n_last)
recon_loss_values = CircularBuffer(n_last)
loss_values = CircularBuffer(n_last)
for i, input_batch in enumerate(train_loader):
input_batch = tuple(data.to(device) for data in input_batch)
#forward
kl_loss, recon_loss = model(input_batch)
loss = kl_weight * kl_loss + recon_loss
#backward
if optimizer is not None:
optimizer.zero_grad()
loss.backward()
clip_grad_norm_(get_optim_params(model),clip_grad)
optimizer.step()
kl_loss_values.add(kl_loss.item())
recon_loss_values.add(recon_loss.item())
loss_values.add(loss.item())
lr = (optimizer.param_groups[0]['lr'] if optimizer is not None else None)
#update train_loader
kl_loss_value = kl_loss_values.mean()
recon_loss_value = recon_loss_values.mean()
loss_value = loss_values.mean()
postfix = [f'loss={loss_value:.5f}',f'(kl={kl_loss_value:.5f}',f'recon={recon_loss_value:.5f})',f'klw={kl_weight:.5f} lr={lr:.5f}']
postfix = {'epoch': epoch,'kl_weight': kl_weight,'lr': lr,'kl_loss': kl_loss_value,'recon_loss': recon_loss_value,'loss': loss_value,'mode': 'Eval' if optimizer is None else 'Train'}
return postfix
def _train(model, train_loader, val_loader=None, logger=None):
optimizer = optim.Adam(get_optim_params(model),lr= lr_start)
lr_annealer = CosineAnnealingLRWithRestart(optimizer)
model.zero_grad()
for epoch in range(n_epoch):
kl_annealer = KLAnnealer(n_epoch)
kl_weight = kl_annealer(epoch)
postfix = _train_epoch(model, epoch,train_loader, kl_weight, optimizer)
lr_annealer.step()
def fit(model, train_data, val_data=None):
logger = Logger() if False is not None else None
train_loader = get_dataloader(model,train_data,shuffle=True)
val_loader = None if val_data is None else get_dataloader(model, val_data, shuffle=False)
_train(model, train_loader, val_loader, logger)
return model
...
to see full code visit above link
Sample from model
For taking a sample from the model we use the encoder and take mean and sigma from the encoder. After taking mean and sigma, we convert this into the decoder dimension and we get sample smiles.
We first feed <”bos”> to the decoder then this decoder tries to give one character at a time. We run this on a loop from 1 to maximum length. Then, from this, we obtain a list of the tensor. Each tensor represent the character of a smile and then by using a tensortostring function we convert obtained tensor to smiles string.
We give the maximum length up to which we generate smiles if it generates less then maximum we pad it. After converting back to string from tensor, we remove this padding.
class sample():
def take_samples(model,n_batch):
n = n_samples
samples = []
with tqdm(total=n_samples, desc='Generating samples') as T:
while n > 0:
current_samples = model.sample(min(n, n_batch), max_len)
samples.extend(current_samples)
n -= len(current_samples)
T.update(len(current_samples))
samples = pd.DataFrame(samples, columns=['SMILES'])
return samples
I used pycharm for writing this code. All above code of model,trainer,data preprocessing,samples are of seperate files. So, finally to run the code we have a file name run.py and the code for running the model is given below.
from trainer import *
from vae_model import VAE
from data import *
from samples import *
model = VAE(vocab,vector).to(device)
fit(model, train_data)
model.eval()
sample = sample.take_samples(model,n_batch)
print(sample)
Hope you liked it. If you want to know more about molecular generation please visit bayes lab github page here you will find more models on molecular generation.
In the next blog we will get an idea about Adversarial Autoencoders.
If you have any doubts please let us know in the comments section.
Thank you
References
I use this GitHub code in this blog to explain VAE. you can see more models in molecular generation in pytorch into this GitHub page.